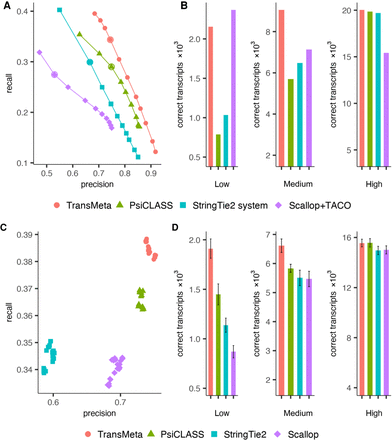
Performance evaluations on the simulated data set S1 under the HISAT2 alignments. (A) Precision-recall curves of the assemblers. The points on the curve of an assembler correspond to the filtering thresholds of the assembler; the circled one, to the default value. (B) Distributions of numbers of correctly recovered transcripts against the tools that were separately counted according to whether their expression is low, medium, or high at the meta-assembly level. (C) The precision and recall of the assemblers on different samples at the individual sample level under their default settings. Different colors correspond to different assemblers, and each point corresponds to a specific sample. (D) Distributions of averaged numbers of correctly recovered transcripts against the tools that were separately counted according to whether their expression is low, medium, or high at the individual sample level. The error bars show the standard deviation across the samples.











