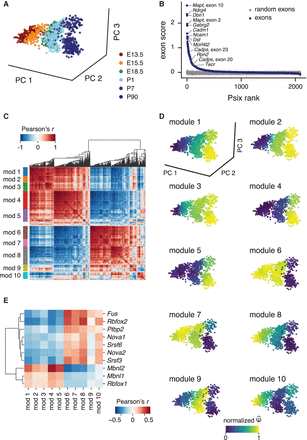
Psix identifies alternative splicing patterns in mouse midbrain development. (A) Mouse midbrain dopamine neurons collected at different stages of development (Tiklová et al. 2019), plotted by the first three principal components of normalized gene expression counts. (B) Psix scores of cassette exons, compared with the scores of randomized exons. Some exons known to be regulated in neurogenesis
are highlighted. (C) Correlation map of the neighbor average 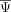 of the cell state–associated exons. Modules were identified with a modification of the UPGMA algorithm. (D) Neighbor average normalized
of the cell state–associated exons. Modules were identified with a modification of the UPGMA algorithm. (D) Neighbor average normalized 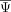 of the exons in each module. (E) Correlation of module splicing with gene expression of splicing factors that were enriched for binding in the cell state–associated
exons identified by Psix. The 10 splicing factors with significant correlation with at least one module (FDR ≤ 0.05, Pearson's
r ≥ 0.25) are shown.
of the exons in each module. (E) Correlation of module splicing with gene expression of splicing factors that were enriched for binding in the cell state–associated
exons identified by Psix. The 10 splicing factors with significant correlation with at least one module (FDR ≤ 0.05, Pearson's
r ≥ 0.25) are shown.











