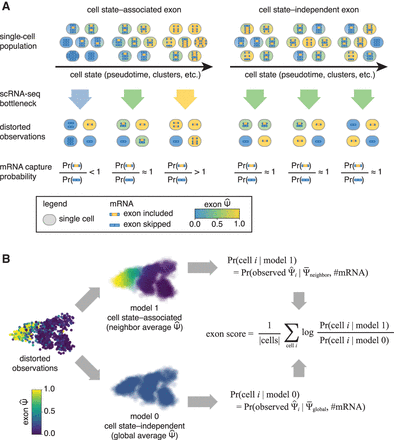
Conceptual model. (A) Cell state–associated exons change across the phenotypic landscape of a single-cell population. Cell state–independent exons
do not change across the phenotypic landscape. Low capture efficiency in scRNA-seq experiments adds additional technical variance
depending on the number of captured mRNA molecules. The probability of capturing each alternative isoform depends on the underlying
distribution of exons in the single-cell population. (B) Psix compares the likelihood of each single-cell observation given two models: model 1, in which the exon is cell state
associated (probability of the cell's 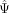 given the average
given the average 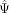 of its k nearest neighbors), versus model 0, in which the exon is cell state independent (probability of the cell's
of its k nearest neighbors), versus model 0, in which the exon is cell state independent (probability of the cell's 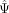 given the average Ψ of all cells in the data set). Model 1 is more likely for a cell state–associated exon. For a cell state–independent
exon, the expected
given the average Ψ of all cells in the data set). Model 1 is more likely for a cell state–associated exon. For a cell state–independent
exon, the expected 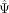 of any cell is the same irrespective of its position in the cell state manifold. As a result, the expected value of the average
of any cell is the same irrespective of its position in the cell state manifold. As a result, the expected value of the average
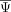 of a neighborhood of cells is the same as the global average
of a neighborhood of cells is the same as the global average 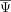 . For this reason, the likelihood of model 1 is similar to the likelihood of model 0 for a cell state–independent exon.
. For this reason, the likelihood of model 1 is similar to the likelihood of model 0 for a cell state–independent exon.











