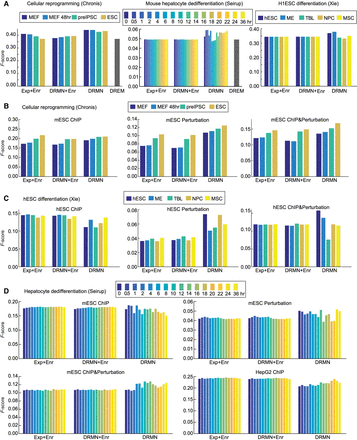
Performance of regulator ranking and inferred networks of DRMN. (A) The F-score of regulator ranking with different strategies for cellular reprogramming, mouse hepatocyte dedifferentiation, and H1ESC differentiation. Each plot shows regulator enrichment of expression-based modules (Exp + Enr), regulator enrichment of DRMN modules (DRMN + Enr), regulator ranking based on inferred DRMN networks (DRMN), and regulator ranking from DREM (when applicable). (B) F-score-based performance of inferred networks and baseline networks for cellular reprogramming, when compared to ChIP-based, perturbation-based, and intersection of the ChIP and perturbation-based gold standards in mESCs. (C) F-score-based performance of inferred networks and baseline networks for H1ESC differentiation, when compared to ChIP-based, perturbation-based, and intersection of the ChIP- and perturbation-based gold standards in hESCs. (D) F-score-based performance of inferred networks and baseline networks for mouse hepatocyte dedifferentiation, when compared to ChIP-based, perturbation-based, and intersection of the ChIP- and perturbation-based gold standards for mESCs, as well as the ChIP-based gold standard from the HepG2 cell line. In each panel, we show the baseline networks from regulator enrichment of expression-based modules (Exp + Enr), regulator enrichment of DRMN modules (DRMN + Enr), and inferred networks with edges scored by input feature multiplied by estimated regression coefficients (DRMN).











