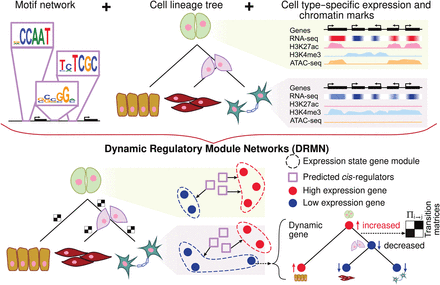
Overview of dynamic regulatory module networks (DRMNs). Inputs are a lineage tree over the cell types, cell type–specific expression levels, a motif-based network, and optionally, cell type–specific features such as histone modification marks or chromatin accessibility signal. The output is a learned DRMN, which consists of cell type–specific expression state modules, their regulatory programs, and transition matrices (white–black matrices) describing the dynamics between the cell types. Expression states of individual genes can be traced on the tree to identify “dynamic” or “transitioning” genes.











