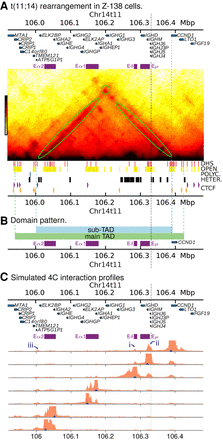
Simulations of the rearranged CCND1 region in Z-138 cells. (A) Simulated Hi-C for Z-138 cells which possess a t(11;14) translocation (hg19_z138 genome build). Positions of the IGH super-enhancers are shown as purple blocks above the map. Simulation input data (DHSs, chromatin states, and CTCF peaks) are shown under the map as in previous figures. The gray dashed line shows the position of the translocation, whereas the gray dotted line shows the position of the CCND1 promoter. (B) Colored blocks show the domain pattern. The green block shows the domain obtained via a TAD-calling algorithm (see Supplemental Methods), and the blue block shows a subTAD region which is visible in the interaction map in A. The subTAD boundary positions are determined by the positions of CTCF sites. Colored dotted lines show where the boundaries lie on the map in A. (C) Simulated 4C from viewpoints positioned at DHSs across the region (blue triangles). Some features are highlighted with numbered arrows as discussed in the text.











