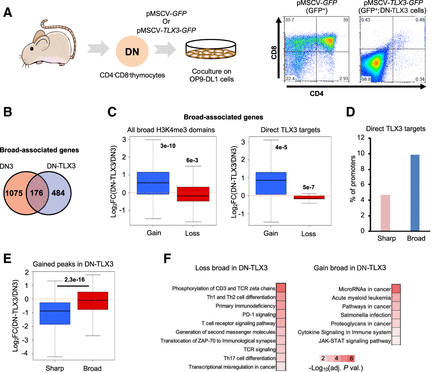
Dynamics of broad H3K4me3 domains in a mouse model of T-ALL. (A) Experimental workflow. Double-negative thymocytes were infected with a retroviral vector expressing TLX3 oncogene and/or the GFP. After 1 mo of coculture with OP9-DL1 stromal cells, the GFP-positive cells were purified and analyzed by FACS. TLX3 expression was confirmed by western blot using a custom-made antibody (Supplemental Figure S10A; Dadi et al. 2012). (B) Overlap between the broad H3K4me3 domains found in DN thymocytes from wild-type mice and TLX3+ leukemic cells (DN-TLX3). (C) Boxplot showing the fold change of expression of genes associated with broad H3K4me3 domains only in DN-TLX3 (gain) or DN (loss; left panel). (Right) Only the direct targets of TLX3 were analyzed. (D) Percentage of broad- and sharp-associated genes in DN-TLX3 cells that are direct targets of TLX3 based on TLX3 ChIP-seq. (E) Two sets of equally expressed genes associated with broad or sharp H3K4me3 peaks in DN-TLX3 cells were defined. The fold change of the expression between DN-TLX3 and DN was determined. (F) KEGG pathways significantly enriched for the gene sets associated with broad H3K4me3 domains only in DN-TLX3 (gain) or DN (loss; adjusted P-value < 0.01). The inverted log10 of the Benjamin-corrected P-value is shown. (C) Significance was assessed by a Wilcoxon signed-rank test. (E) Significance was assessed by a Wilcoxon rank-sum test.











