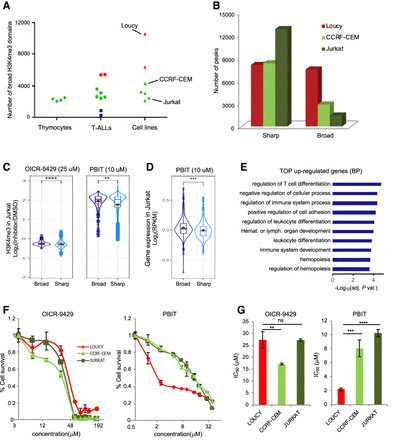
Extent of broad H3K4me3 domains and differential drug sensitivity. (A) Number of broad H3K4me3 domains in the thymic T cell populations, the primary T-ALL, and the T-ALL cell lines. (B) Number of sharp and broad-H3K4me3 peaks in the indicated cell lines. (C) Box plots showing the distribution of the observed fold change of H3K4me3 ChIP-seq signals at broad and sharp promoters in Jurkat cells treated with the indicated inhibitor or DMSO. (D) Box plots showing the distribution of the observed fold change of RNA-seq FPKM at broad- and sharp-associated genes in Jurkat cells treated with PBIT or DMSO. (E) Top 10 GO terms for biological process enriched at genes associated with broad H3K4me3 domains that were up-regulated after PBIT treatment of Jurkat cells. (F) Proliferation assay of Loucy, CCRF-CEM, and Jurkat T-ALL cell lines in the presence of the indicated concentrations of OICR-9429 or PBIT inhibitor for 72 h. Results are the mean of three independent experiments. (G) Bar chart shows average ± SD IC50 values (n = 3) obtained in the proliferation assays shown in F. Significance in C and D was assessed by the Wilcoxon rank-sum test. Significance in G was assessed by a two-sided Student t-test. Error bars, SD. (****) P < 0.0001, (***) P < 0.001, (**) P < 0.01.











