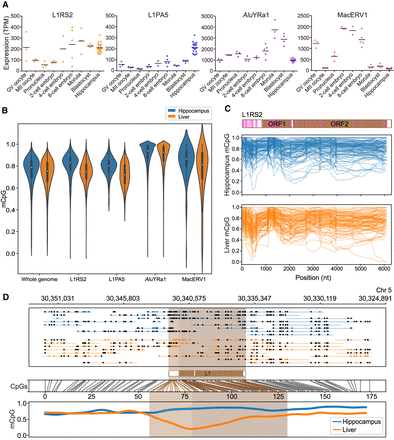
Genome-wide analyses of young TE subfamily transcription and methylation. (A) Subfamily-wide TE expression measured by RNA-seq (TPM) and an existing strategy to account for multimapping reads (Faulkner et al. 2008, 2009; Hashimoto et al. 2009). Data were obtained from prior studies (Wang et al. 2017; Yin et al. 2020) and encompassed GV and MII oocytes, early embryonic development, and adult hippocampus. Horizontal bars represent the mean of biological replicates. (B) Violin plots showing CpG methylation ascertained by ONT sequencing upon animal ON22213 hippocampus and liver. Results are shown for the whole genome (6-kbp windows), L1RS2 and L1PA5 copies >6 kbp, AluYRa1 copies >300 bp, and MacERV1 long terminal repeats >300 bp. (C) Composite L1RS subfamily methylation profiles. Each graph displays 100 profiles. A schematic of the L1RS2 consensus sequence is provided at top, with CpG positions indicated by pink bars. (D) Exemplar methylation profile of an L1RS2 element located on Chromosome 5 and hypomethylated in the liver. The panel is composed as described for Figure 3A.











