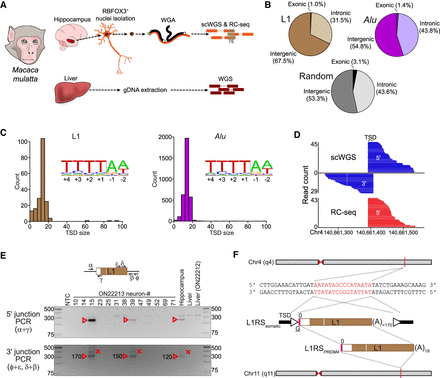
Characterization of germline and somatic macaque TE insertions. (A) Genomics experimental design. Individual hippocampal neuron (RBFOX3+) nuclei from two rhesus macaques (ON22212 and ON22213) were subjected to whole-genome amplification (WGA), followed by Illumina scWGS and RC-seq, to identify somatic TE insertions. Bulk liver DNA was analyzed with Illumina WGS to discriminate germline and somatic variants. (B) Percentages of exonic, intronic, and intergenic nonreference L1 (top left) and Alu (top right) insertions. Genomic features were annotated according to RefSeq coordinates, with the underlying proportions of each feature (random expectation) shown at bottom. (C) Target site duplication (TSD) size distributions for nonreference L1 (left) and Alu (right) insertions, as annotated by TEBreak. Inset sequence logos (Crooks et al. 2004) display the observed integration site nucleotide composition for each TE family. These resembled the L1 endonuclease motif. (D) A somatic L1RS2 insertion (L1RSsomatic) was detected on Chromosome 4 of animal ON22213 hippocampal neuron #15. Reads spanning the 5′ or 3′ L1–genome junctions of this event are shown, as is the corresponding TSD. (E) PCR validation of L1RSsomatic. Primer (symbols α, ε, δ, γ, β, and Φ) positions relative to the L1 insertion are indicated in the schematic provided at top. The 5′ L1–genome junction was amplified by combining primers α and γ, whereas nested PCR (ε + Φ then δ + β) was used to amplify the 3′ L1–genome junction. Reaction input in each case consisted of nontemplate control (NTC), 13 ON22213 hippocampal neurons analyzed with scWGS and RC-seq, bulk ON22213 hippocampus and liver DNA, and bulk ON22212 liver. Red arrowheads and crosses indicate amplicons confirmed as on-target and off-target, respectively, by capillary sequencing. Numbers next to confirmed 3′ L1–genome junction bands indicate the L1 poly(A) tract length for that amplicon. (F) Complete sequence characterization of L1RSsomatic. TSD nucleotides are highlighted in red. The intergenic L1 was full length (L1RS2 subfamily consensus start position 0), carried a 4-bp 5′ transduction (pink rectangle) with an untemplated guanine (underlined G), and was followed by a long, pure 3′ poly(A) tract. The transduction indicated a putative donor L1 intronic to the PRDM4 gene on Chromosome 11 (L1RSPRDM4).











