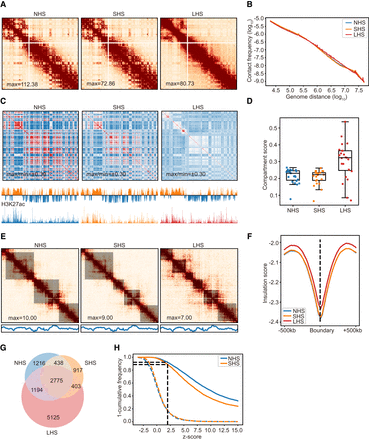
Chromatin conformation remained stable after short-term HS (SHS), whereas it is mostly altered after long-term HS (LHS). (A) Chr 6 is shown with 500-kb resolution as an example of the contact maps in K562 cells during the process of HS. (B) Contact frequency decay curves at normal HS (NHS), SHS, and LHS conditions. (C) Chromatin compartmentalization at the three conditions. Chr 6 is shown as an example. The autocorrelation matrices together with the first eigenvector profiles are shown. In the first eigenvector, compartment B is colored as blue and A as orange. (D) Genome-wide compartment scores at each condition. (E) TADs detected in a 4-Mb region centered at the TSS of HSPA1A gene are shown as an example, together with the corresponding contact maps. The insulation score profiles of the same region are also shown below. The detected TADs are shaded. (F) Genome-wide insulation score profiles around TAD boundaries at the three conditions. (G) Numbers and proportions of overlap of chromatin loops at the three conditions. (H) Distributions of z-scores of LHS-specific loops in NHS and SHS conditions. Dashed lines indicate the distributions of z-scores of the randomly chosen bin pairs with the same genome distances. Black dashed line shows the proportion of LHS-specific loops that had z-scores of contacts greater than two compared with their flanking regions.











