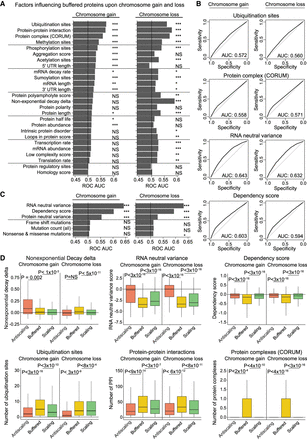
Multiple genetic and biochemical factors predict protein buffering upon aneuploidy. (A) Bar graphs displaying ROC area under the curve values for each independent factor upon chromosome gain or loss. Genes classified as “buffered” upon chromosome gain or loss were set as the true positive fraction. Significance was calculated by performing 10,000 random permutations and bootstrapping P-values, where (*) indicates < 0.05, (**) < 0.005, and (***) < 0.0005. (B) ROC curves are displayed for certain key factors. Genes classified as “buffered” upon chromosome gain or loss were set as the true positive fraction. (C) ROC AUC values for data set–specific factors upon chromosome gain or loss. Genes classified as “buffered” upon chromosome gain or loss were set as the true positive fraction. (D) Boxplots displaying buffering factor scores, per difference category upon chromosome gain or loss. Six buffering factors are displayed: nonexponential decay delta, RNA neutral ploidy variance, dependency score, number of ubiquitination sites, number of protein–protein interactions, and number of protein complexes. P-values are from two-sided t-tests. Boxplots display the 25th, 50th, and 75th percentiles of the data, and the whiskers indicate 1.5 interquartile ranges.











