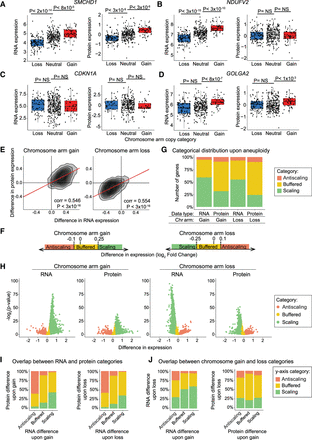
Protein expression differences are frequently buffered upon chromosome gain or loss. (A–D) Normalized RNA and protein expression levels of the indicated genes (SMCHD1, NDUFV2, CDKN1A, and GOLGA2) are displayed for cell lines in which the chromosome that that gene is encoded on is either lost, neutral, or gained. Boxplots display the 25th, 50th, and 75th percentiles of the data, and the whiskers indicate a 1.5 interquartile range. P-values were calculated using two-sided t-tests. (E) A density plot comparing the difference in RNA expression versus the difference in protein expression levels upon the gain (left) or loss (right) of the chromosome arm the gene is located on. The Pearson correlation coefficient and P-value are displayed. Linear regressions (red) with 95% confidence intervals are displayed. (F) Diagram displaying the categorical cutoff points for gene difference in expression. The cutoffs are −0.1 and 0.25 for chromosome gain and −0.25 and 0.1 for chromosome loss. The categorical cutoffs are labeled with dashed lines. (G) The percentage of RNAs and proteins that fall into each difference category upon chromosome gain and loss are displayed. (H) Volcano plots displaying the difference in RNA expression or protein expression upon chromosome arm gain (left) or chromosome arm loss (right) versus the P-value for each gene. Genes are color-coded based on a categorical distribution as either scaling, buffered, or antiscaling. (I) Bar graphs displaying the percentage of genes in each RNA difference category (x-axis) whose corresponding proteins fall into each of the indicated expression categories upon chromosome gain (left) and loss (right). (J) Bar graphs displaying the percentage of RNAs in each RNA difference category upon chromosome gain (x-axis) that fall into each of the indicated expression categories upon chromosome loss (y-axis; left). Percentage of proteins, per protein difference category upon chromosome gain (x-axis), that fall into each expression category upon chromosome loss (y-axis; right).











