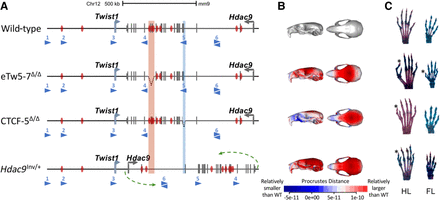
Alteration of Twist1 regulatory elements in mice. (A) Scheme of the aberrations within the Hdac9 sequence. In the eTw5-7Δ/Δ model, a 23-kb sequence, containing Hdac9 exons 20–23 and Twist1 enhancers (eTw5-7), was deleted (highlighted red rectangle). In the CTCF-5Δ/Δ model, a 1.5-kb sequence containing Hdac9 exons 6–7 was deleted (highlighted blue rectangle). In the Hdac9INV/+ model, the whole Hdac9 sequence, 890 kb long, was inverted. The inversion breakpoints are marked by green arrows (mm9, Chr 12: 34,721,220–35,613,000). (B) Heat maps showing anatomical distributions of shape change compared to the wild type for eTw5-7Δ/Δ, CTCF-5Δ/Δ, and Hdac9INV/+ mice, demonstrated by side view (right) and superior view (left) (red is larger and blue is smaller compared to the grand mean). (C) Polydactyly was found in both hindlimb (HL) (71%) and forelimb (FL) (50%) of eTw5-7Δ/Δ mice, whereas polydactyly was found only in the HL of CTCF-5Δ/Δ (32%) and Hdac9INV/+ (8%) mice. The polydactyly is marked by asterisks.











