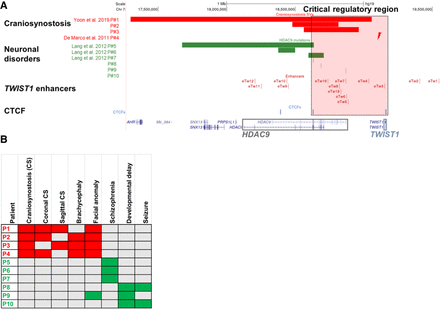
Structural variants containing HDAC9 (but not TWIST1) in patients with craniosynostosis and neuronal disorders. (A) SVs encompassing HDAC9 but not TWIST1 indicate the potential location of a critical TWIST1 regulatory region (highlighted by the dashed light pink rectangle). Red bars represent three craniosynostosis patients with de novo HDAC9 deletions (P1–P3), and red lightning represents the translocation t(7;12)(p21.2;p12.3) breakpoint located between HDAC9 and TWIST1 in craniosynostosis patient (P4). Green bars represent three schizophrenia patients (P5–P7) with de novo HDAC9 deletions, and green lines represent three patients with neurological phenotypes (P8–P10) with SNVs in HDAC9. Blue lines represent CTCF sites involved in chromatin looping in the HDAC9-TWIST1 locus. Red lines represent TWIST1 enhancers located in introns or exons of the HDAC9 sequence and intergenic regions. (B) Human Phenotype Ontology heat map of patients’ common clinical features. Gray boxes represent either absent or unreported symptoms.











