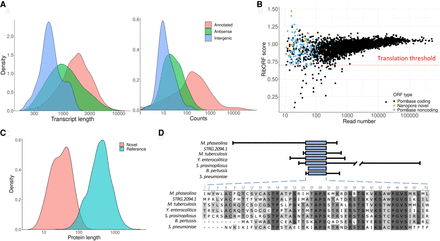
New transcripts and peptides. (A) Transcript length and gene expression for novel antisense and intergenic transcripts. Novel antisense transcripts tend to be longer than intergenic transcripts, and both classes of transcripts are shorter than already characterized ones (Wilcoxon test, P-value < 10−5). Gene expression levels are lower in novel transcripts compared with not novel ones (Wilcoxon test, P-value < 10−5). Novel antisense and intergenic transcripts show a similar expression level distribution. (TPM) Transcripts per million, quantification provided by StringTie. (B) Prediction of translated ORFs. The plot shows the RibORF score versus the number of Ribo-seq mapped reads for different classes of transcripts. ORFs with at least 10 mapped reads and a RibORF score higher than 0.7 were selected as translated. The score is based on the Ribo-seq read 3-nt periodicity and homogeneity. Nanopore novel indicates transcripts that did not overlap any annotated transcript, 12 of these transcripts contained ORFs with translation signatures. PomBase noncoding indicates annotated ncRNAs that also had translation signatures. (C) Novel translated ORFs are shorter than annotated ones. Comparison of aa length for ORFs with evidence of translation in novel transcripts and annotated coding sequences. Differences are statistically significant (Wilcoxon test, P-value < 10−5). (D) New protein identified in S. pombe. The protein labeled as STRG.2094.1 showed significant similarity to other bacteria proteins (BLASTP e-values between 10−2 and 10−7) and, in eukaryotes, only to a protein from the fungus Macrophomina phaseolina (e-value <10−9). The blue box represents the homologous region; lines at the side represent additional protein sequence.











