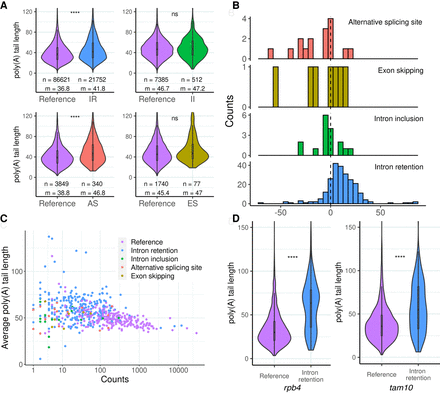
Poly(A) length in alternative transcript isoforms. (A) Distribution of poly(A) length by isoform type. We computed poly(A) length for all dRNA reads with nanopolish. Only reads with the label PASS were considered. (n) Number of reads with poly(A) length information; (m) median poly(A) length. (IR) Intron retention; (II) intron inclusion; (AS) alternative splicing site; and (ES) exon skipping. Significant differences were identified for isoform retention and AS site events compared with the corresponding reference isoforms. (***) P-value < 10−5, Wilcoxon test. (B) Difference in the average poly(A) length between the alternative and reference isoforms. Longer poly(A) lengths were consistently observed for IR isoforms. (C) Negative correlation between average poly(A) length and expression level for reference and alternative transcript isoforms. The data are only for genes in which we detected alternative isoforms. Reference refers to the annotated isoform. Spearman's ρ = −0.41, P = 3.14 × 10−23. (D) Examples poly(A) length differences between the reference and the IR isoforms. We computed the poly(A) length for the dRNA reads that correspond to each of the isoforms. The first example corresponds to RNA polymerase II subunit 4 (rpb4, SPBC337.14), with ∼27% of the reads corresponding to the IR isoform. The second example corresponds to a nucleolar RNA-binding protein also implicated in mRNA processing (tam10, SPBC14C8.19), with ∼18% of the reads mapping to the IR isoform. In both cases poly(A) length showed a significant tendency to be longer in the IR isoform. (***) P-value < 10−5; (ns) nonsignificant, Wilcoxon test.











