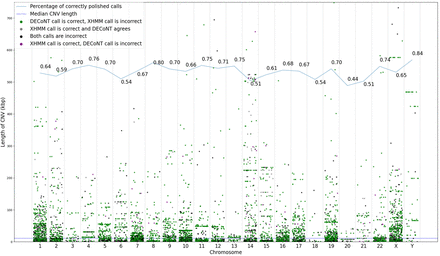
Each dot in this figure corresponds to one of the 6832 CNV calls made by XHMM on our test data set (the 1000 Genomes Project WES data set test samples). The ground truth is the CNV calls made by CNVnator on the corresponding WGS samples. The calls are stratified w.r.t. their chromosomes. The y-axis of the plot represents the length of calls. A gray dot indicates that the XHMM call is not changed by DECoNT and the prediction matches the ground truth (correct), whereas a black dot indicates that both DECoNT's and XHMM's decisions do not match the ground truth (incorrect). A green dot indicates the call is corrected by DECoNT and the XHMM call was incorrect. Finally, a purple dot indicates that DECoNT changes the prediction of XHMM and both the original and the changed calls are incorrect. For each chromosome, a random jitter is added to the x-axis for better visualization. The solid line on the top of the figure shows the ratio of the number of DECoNT-corrected calls and the number of all XHMM calls in that chromosome. The dashed line indicates the median CNV call length across chromosomes. For each polished tool, we used 90% of the calls made on 802 1000 Genomes Project samples for training and the remaining 10% of the calls for testing. This roughly corresponds to a test set size of 80 samples.











