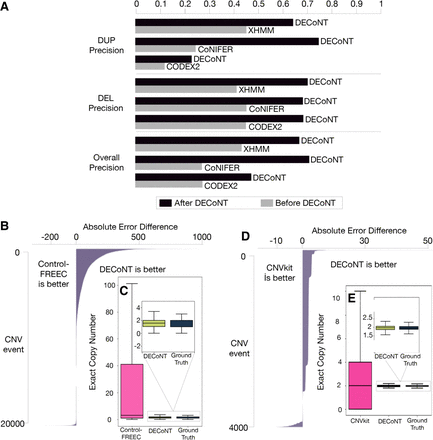
The performance comparison of the WES-based CNV callers before and after polishing with DECoNT. (A) For the tools that predict the existence of a CNV event (XHMM, CoNIFER and CODEX2) are evaluated with respect to duplication call precision, deletion call precision, and overall precision, DECoNT improves the performance for all tools in all settings and results in substantial improvements. Different shades of gray represent different tools, and the attached black bars represent the DECoNT-polished version of those tools. (B) In this panel, we compared Control-FREEC and the DECoNT-polished with respect to absolute error (AE) difference on each sample (i.e., events). Bars to the right indicate the magnitude of the improvement owing to polishing of DECoNT. For more than half of the samples, DECoNT results show improvement. (C) The distribution of the unpolished Control-FREEC predictions in the test samples (pink) is quite different than the ground-truth CNV distribution. On the other hand, DECoNT polished versions of the same events (dark blue) highly resemble the distribution of the ground-truth calls. Black lines across the boxes are median lines for the distributions. Black vertical lines are whiskers and 1.5× inter-quartile range is defined with the horizontal lines at the top and bottom of the whiskers. The 1000 Genomes Project WGS samples are used as ground-truth calls in all analyses. (D,E) The results for CNVkit, similar to B and C. For each polished tool, we used 90% of the calls made on 802 1000 Genomes Project samples for training and the remaining 10% of the calls for testing. This roughly corresponds to a test set size of 80 samples.











