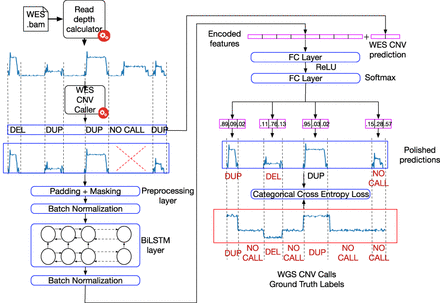
Learning workflow of DECoNT. First, BAM file that corresponds to a WES data set from the 1000 Genomes Project is used to calculate exome-wide read depth, which is input into a third-party WES-based CNV caller. The caller generates the calls for various regions that could be (1) a binary prediction like duplication, deletion (e.g., XHMM) (Fromer et al. 2012) as shown in the figure, or (2) an integer value that indicates the exact copy number (i.e., Control-FREEC) (Boeva et al. 2012). The read depth of the regions for which a call has been made is input to a Bi-LSTM model. Encoded features are passed from a series of fully connected (FC) layers along with the original prediction of the caller algorithm. Using the ground-truth calls from the WGS data of the same sample, the method learns to predict (correct) the calls using cross-entropy loss for the binary outputs (as shown in the figure) and using mean-squared loss for integral calls.











