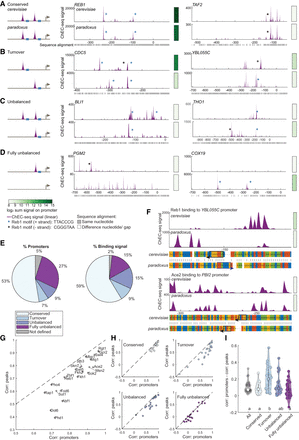
Promoter evolution and binding-site turnover. (A–D) Four evolutionary classes of TF binding variation. Schemes are shown in the left panels; genome browser snapshots of Reb1-bound promoters are shown as examples in the middle and right panels. Sequence alignment between orthologs is presented below each example, x-axis represents the location on the promoter relative to TSS (figure legend below D). (A) Conserved: all binding sites are species-conserved. (B) Turnover: reciprocal gain and loss of binding sites. (C) Unbalanced: species-specific sites along with conserved sites. (D) Fully unbalanced: binding sites appear in only one ortholog. (E) Proportion of number of promoters and binding signal per promoter class. Shown is the distribution of the full set of TFs; proportion per TF is presented in Supplemental Figure S13A. Binding signal refers to the ChEC-seq signal in the higher-bound ortholog, normalized by the total signal in that ortholog. (F) Examples of short-distance binding-site turnover. Shown are the binding signal in both alleles, and the sequence alignment in the lower panel. Boxes mark motifs, and arrows mark the motif's strand. (G) Correlation on promoters is higher than correlation on peaks when comparing orthologs. Shown are correlation coefficients between orthologs, over motif-associated peaks (y-axis) and promoters (x-axis), among all promoters. Here, we summed the binding signal only on peaks within peak-containing promoters, but the correlation coefficients were quantitatively similar to those obtained from the more simplistic approach of summing up the signal over the full promoter, as presented in Figure 1B (cf. Fig. S15A). (H) The shift in promoter correlation versus peak correlation is more apparent at turnover and unbalanced promoters. Shown is the correlation between orthologs, summing over promoters (x-axis) and over motif-associated peaks (y-axis) as in G, per promoter class. (I) Turnover promoters show a higher promoter similarity despite lower peak similarity. Shown are the differences between correlation on promoters to correlation on peaks for the different promoter classes. Each dot represents a TF; letters represent statistically distinguished groups after Tukey's honestly significant difference test.











