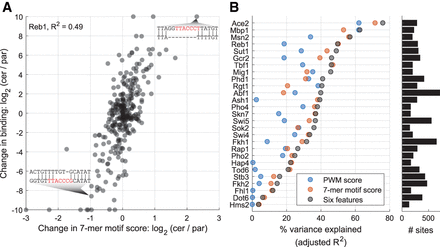
Sequence variation predicts DNA binding variation. (A) Change in motif score predicts variation in Reb1 binding. Shown is the log2-ratio of 7-mer motif scores (x-axis) and ChEC-seq signal (y-axis) between S. cerevisiae and S. paradoxus orthologs, at peaks associated with a nonconserved Reb1 motif (i.e., a different motif sequence appears in each ortholog). Sequence alignment at two specific sites is presented: S. cerevisiae ortholog on the upper row, S. paradoxus ortholog in the lower row, and the Reb1 motif written in red. (B) Linear models predict binding variation at peaks associated with nonconserved motifs. Shown is the percentage of explained variability (R2× 100) for each TF, using three models: in vitro PWM score, 7-mer motif score derived from our data, and a compilation of these with another four predictors (see text). Predictions for other peak categories are presented in Supplemental Figure S11A.











