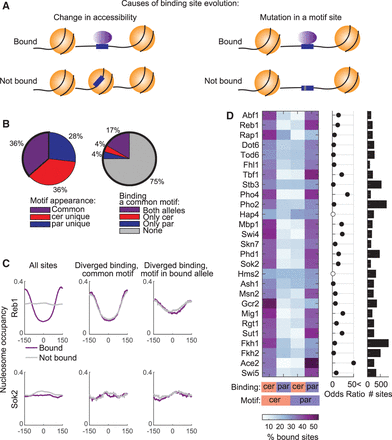
Differential TF binding to the two alleles correlates with variations in motif sequence, but differences in motif accessibility play a minor role. (A) Suggested mechanisms for TF binding evolution: (left) motif site is conserved, but in the unbound allele it is occupied by a nucleosome and therefore not accessible for TF binding; and (right) motif site is lost owing to a single-nucleotide variation. Nucleosomes are illustrated in orange; TF in purple oval; motif site in blue box; nucleotide variation as a gray stripe. (B) Proportion of motif sites (left) and proportion of bound sites among the common motif sites (right). (Left) Proportion of motif sites that are common to both orthologs, and sites that appear only in a certain ortholog (cer- or par-unique) among all in vitro defined motif sites of the full set of 27 TFs (62,970 are common, 63,455 cer-unique, 46,440 par-unique) (for TF-specific proportions, see Supplemental Fig. S6). (Right) Proportion of binding to common motif sites. (C) Nucleosome occupancy does not explain differential binding between orthologs. Presented are nucleosome occupancy profiles averaged over motif sites, centered at the binding motif of Reb1 (upper) and Sok2 (lower). (Left) All motif-containing sites, divided into bound sites (purple) and nonbound sites (gray). (Middle) Common motif sites that show diverged binding. Nucleosomes at the bound allele are in purple, and nucleosomes at the nonbound allele are in gray. (Right) Unique motif sites with diverged binding. Nucleosomes at the bound allele, which harbors a motif, are in purple, and nucleosomes at the nonbound and motifless allele are in gray (for profiles of all TFs, see Supplemental Fig. S7). (D) Binding to unique motif sites, with biased binding to the motif-containing allele. (Left) Percent of bound sites. (Middle) Odds ratio of Fisher's exact test; full black circles indicate significant comparisons (P-value < 0.05, FDR corrected). (Right) Number of unique motif sites.











