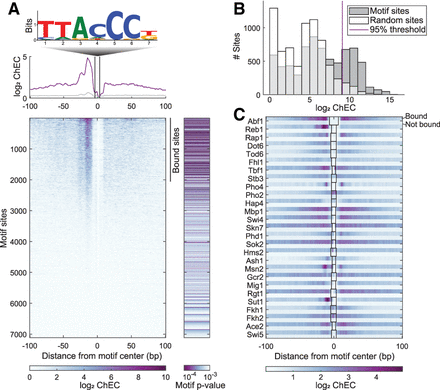
Transcription factors bind a select subset of motif-containing sites. (A) Reb1 binding signal to motif-containing sites (potential binding sites). (Top) In vitro motif of Reb1 (Fordyce et al. 2010). (Middle) Average ChEC-seq signal (5′ end of reads) in a logarithmic scale; bound sites presented in purple, nonbound sites presented in gray. (Bottom left) Heatmap of ChEC-seq signal at 7115 sites that contain the Reb1 motif in both hybrid genomes; (bottom right) motif P-value according to FIMO (Grant et al. 2011). (B) Binding level distribution of Reb1 in Reb1 motif sites and in random sites. Binding level threshold was set as the 95% of random site distribution. This threshold defines the bound sites indicated in A. (C) Signal around motif sites of all examined TFs, at bound (top row) and nonbound (bottom row) sites. Boxes indicate motif size. Full profiles are presented in Supplemental Figure S2.











