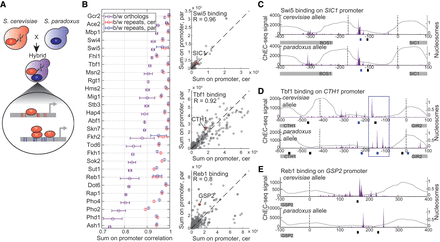
Experimental system to profile cis variation in transcription factor binding. (A) Scheme of the experimental system. S. cerevisiae strains where a TF was fused to a MNase (illustrated as scissors) were crossed with a WT S. paradoxus strain to form a hybrid, on which the ChEC-seq method was applied to profile in vivo TF binding. Orthologous promoters harbor sequence variations (red and blue lines) and differential binding levels. (B) Global similarity in orthologous binding of the 27 TFs examined here. Presented is the Pearson correlation coefficient of sum of signal on all yeast promoters (6701 promoters), between experimental replicates (S. cerevisiae genome in red, S. paradoxus genome in blue), and between orthologous genomes (purple). Data are the mean and standard deviation of two to five repeats. (Right) Promoter binding of three TFs; each point is the sum of signal on a specific gene promoter. (C–E) Examples for TF binding to orthologous promoters. (C) Conservation of Swi5 binding to SIC1 promoter, S. cerevisiae ortholog (upper) and S. paradoxus ortholog (lower). ChEC-seq signal is the 5′ end of reads, presented in purple. Nucleosome occupancy data of the hybrid (Tirosh et al. 2010) are presented as gray lines. Transcription start sites are presented in gray dashed lines (Pelechano et al. 2013; Park et al. 2014). For Swi5, CCAGC motif sequences are marked in blue (plus strand) and black (minus strand) boxes. ORFs are presented as gray boxes. (D) Binding-site turnover of Tbf1 to GIR2/CTH1 promoter. The blue box marks the region of binding-site turnover, in which the Tbf1 motif appears on the plus strand in the S. cerevisiae allele (ACCTA), and the same motif realization appears on the minus strand in the S. paradoxus allele (TAGGT), where motif sequences partially overlap. Consensus motif of Tbf1 is [C/A]CCTA. (E) Divergence in Reb1 binding to GSP2 promoter. Annotation as in D; Reb1 consensus motif is TTACCC[G/T].











