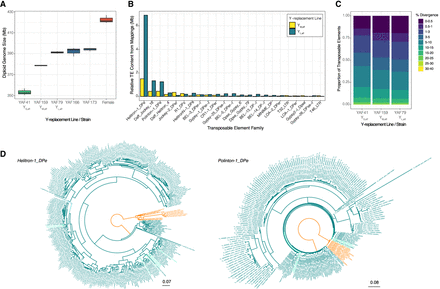
Y Chromosome expansions in D. affinis by the same TEs as in D. pseudoobscura. (A) Diploid genome size estimations from D. affinis Y-replacement lines. Rightmost boxplot (red) is the backcross female strain. (B) TE abundance in YM, aff and YL, aff relative to YS, aff from paired-end mappings to the TE library, made in a similar manner as in Figure 3B. TEs for which the absolute difference is >50 kb are shown. (C) Divergence in TE copies from whole-genome Illumina sequencing across the Y's estimated by dnaPipeTE, made in a similar manner as in Figure 3C. (D) Phylogenetic trees of Helitron-1_DPe (left) and Polinton-1_DPe (right) found in D. pseudoobscura (green), D. affinis (orange), and D. pseudoobscura YL (teal). Repbase sequences are highlighted in blue.











