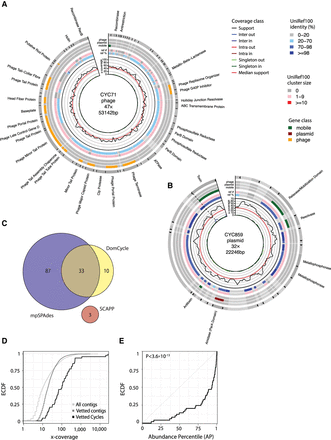
ecMGEs in the gut of a healthy adult. (A) A putative gut phage. Shown from inner to outer circles are nucleotide-level coverage profiles, UniRef100 hits (sequence identity [AAI] to the best UniRef100 hit and the number of UniProt genes in the UniRef100 cluster), and gene classification. Gene descriptions for select phage-associated genes are specified outside. Cycles are cut open at the start of their linear sequence for visualization purposes. (B) Same as A, for a putative plasmid. (C) Genomes corresponding to reported cycles pairwise aligned and clustered using a threshold ANI of 95%. Shown is cluster breakdown for DomCycle, metaplasmidSPAdes, and SCAPP. (D) Shown are the empirical cumulative distribution functions (ECDFs) for the AMC of the 49 vetted dominant cycles that correspond to putative ecMGEs (black), the background AMC defined as the AMC of contigs vetted in the same way as dominant cycles except for the circularity condition (dark gray), and the background coverage all contigs in the assembly (light gray). (E) Shown for all 49 vetted dominant cycles is the ECDF of the cycle abundance percentile, defined as the percentile of the cycle AMC within the background AMC distribution. P-value computed using a one-sided Kolmogorov–Smirnov test is shown.











