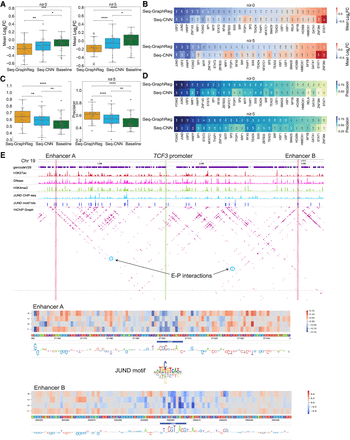
Seq-GraphReg accurately predicts the regulatory effects of transcription factor knockouts by in silico motif ablation. (A) Distributions of true mean logFC (over 100 predicted target genes) over all TFs for Seq-GraphReg, Seq-CNN, and baseline for n ≥ 0 and n ≥ 5, where n denotes the number of enhancer–promoter (E–P) interactions. The median of true mean logFCs for predicted genes by Seq-GraphReg is negative, and the distribution is significantly more down-regulated than that of Seq-CNN and baseline (Wilcoxon signed-rank test). (B) Heatmaps of the true mean logFC of the top 100 predicted genes by Seq-GraphReg and Seq-CNN for each TF, for n ≥ 0 and n ≥ 5. For the majority of TFs, the mean logFC of Seq-GraphReg's predicted targets is more negative than that of Seq-CNN's targets. (C) Distributions of precision values (fraction of true significantly down-regulated genes among 100 predicted genes) of all TFs for Seq-GraphReg, Seq-CNN, and baseline, for n ≥ 0 and n ≥ 5. The precision is always highest in Seq-GraphReg and significantly greater than for Seq-CNN and baseline (Wilcoxon signed-rank test). (D) Heatmaps of precision values (fraction of true significantly down-regulated genes among the top 100 predicted genes) for Seq-GraphReg and Seq-CNN for each TF and for n ≥ 0 and n ≥ 5. For the majority of TFs, the precision of Seq-GraphReg is higher than Seq-CNN. (E) A visual example of the effect of JUND KO on the gene TCF3. JUND motif hits around TCF3 are plotted in blue bars. The promoter of TCF3 is indicated by green lines, and two distal enhancers A (1.08 Mb downstream) and B (1.29 Mb upstream) of the gene TCF3 by red lines. The interactions of enhancers A and B with the promoter of TCF3 in HiChIP graph are marked by blue circles. In silico mutagenesis (ISM) is performed in 100-bp regions of enhancers A and B, each centered at a JUND motif, and the ISM heatmaps are shown. The heatmaps show the difference in predictions (mutated − reference) after applying a mutation at each nucleotide. The heatmaps around the JUND motif in both enhancers A and B are blueish, indicating the importance of JUND motif in these regions for TCF3 expression prediction. The base-level representations of ISM scores are the negative summation of all four scores (only three are non-zero) at each nucleotide.











