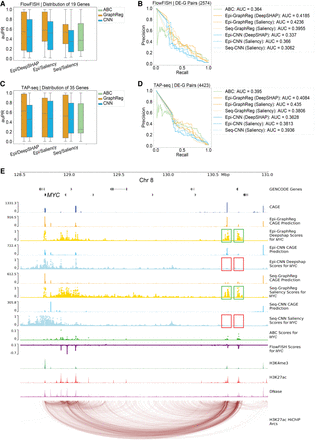
GraphReg models more accurately identify functional enhancers of genes. (A) Distribution of the area under the precision-recall curve (auPR) for 19 genes in K562 cells based on CRISPRi-FlowFISH data. GraphReg models have higher median of auPR than both CNN and activity-by-contact (ABC) models. (B) Precision-recall curves of the GraphReg, CNN, and ABC models for identifying enhancers of 19 genes screened by CRISPRi-FlowFISH. (C) Distribution of auPR for 35 genes in K562 cells based on TAP-seq data. GraphReg models have higher median of auPR than both CNN and ABC models. (D) Precision-recall curves for the GraphReg, CNN, and ABC models for identifying functional enhancers of 35 genes as determined by TAP-seq. (E) MYC locus (2.5 Mb) on Chr 8 with epigenomic data, true CAGE, predicted CAGE using GraphReg and CNN models, HiChIP interaction graph, and the saliency maps of the GraphReg and CNN models, all in K562 cells. Experimental CRISPRi-FlowFISH results and ABC values are also shown for MYC. Feature attribution shows that GraphReg models exploit HiChIP interaction graphs to find the distal enhancers, whereas CNN models find only promoter-proximal enhancers. Green and red boxes show true positives and false negatives, respectively. CNN models miss the distal enhancers and consequently lead to false negatives in very distal regions.











