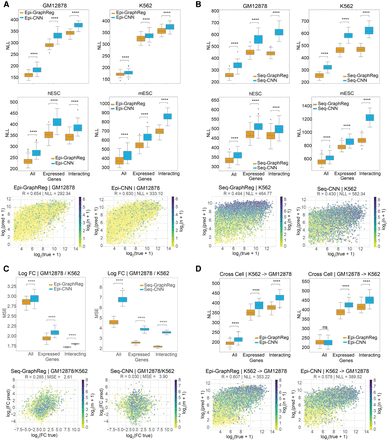
GraphReg models outperform their CNN counterparts for gene expression prediction. (A,B) Negative log-likelihood (NLL, lower is better) between true and predicted CAGE signals of epigenome-based (A) and sequence-based (B) GraphReg and CNN models over 50 random selections of 2000 predicted genes from test chromosomes concatenated from 10 cross-validation experiments with different training, test, and validation chromosomes. Box plots show the distributions of NLL in GM12878, K562, hESC, and mESC for three gene sets: all genes, expressed genes (CAGE signal ≥ 5), and expressed genes with at least one 3D interaction (interacting). The 3D data used in Epi-GraphReg (A) for each cell type is as follows: Hi-C (FDR = 0.001) for GM12878, H3K27ac HiChIP (FDR = 0.01) for K562, Micro-C (FDR = 0.1) for hESC, and H3K27ac HiChIP (FDR = 0.1) for mESC. The 3D data used for Seq-GraphReg (B) is H3K27ac HiChIP (FDR = 0.1) for GM12878, K562, and mESC, and Micro-C (FDR = 0.1) for hESC. Example scatterplots of all predicted test genes that are expressed (CAGE ≥ 5) are plotted for GM12878 in epigenome-based models (A) and K562 in sequence-based models (B), where the genes are color-coded by the number of 3D interactions n. The sequenced-based models have been trained separately (and not using dilated CNN) for K562 and end-to-end for GM12878, hESC, and mESC. (C) Box plots show mean squared error (MSE) of the true and predicted log-fold gene expression changes between GM12878 and K562 in 50 random selections of 2000 predicted genes from test chromosomes concatenated from 10 cross-validation experiments with different training, test, and validation chromosomes. The sets All, Expressed, and Interacting denote the intersections of such sets in GM12878 and K562. Both Epi- and Seq-GraphReg models have better prediction accuracy than their CNN counterparts. The scatterplots of the true log-fold gene expression changes and the log-fold changes derived from the predicted CAGE values by Seq-GraphReg and Seq-CNN, between GM12878 and K562, are shown for expressed genes (CAGE ≥ 5 in both K562 and GM12878). TSS bins are color-coded by the minimum number of 3D interactions in GM12878 and K562 (m). Seq-GraphReg has higher R and lower MSE than Seq-CNN. (D) Epi-GraphReg models show higher cell-to-cell generalization capability than Epi-CNN models. Box plots show the distributions of NLL on the test cell type (K562 or GM12878) when trained on the other cell type over 50 random selections of 2000 predicted genes from test chromosomes of the test cell concatenated from 10 cross-validation experiments with different training, test, and validation chromosomes in the training cell. The models are evaluated on the same test chromosomes in the unseen test cell. HiChIP (FDR = 0.1) is used for both cells. The generalization of Epi-GraphReg from K562 to GM12878 is significantly better (P < 10−4, Wilcoxon signed-rank test) than Epi-CNN in all gene sets. The generalization of Epi-GraphReg from GM12878 to K562 is significantly better (P < 10−4, Wilcoxon signed-rank test) than Epi-CNN in expressed and interacting genes. The scatterplots of all predicted test genes that are expressed (CAGE ≥ 5) are plotted when trained on K562 and tested on GM12878.











