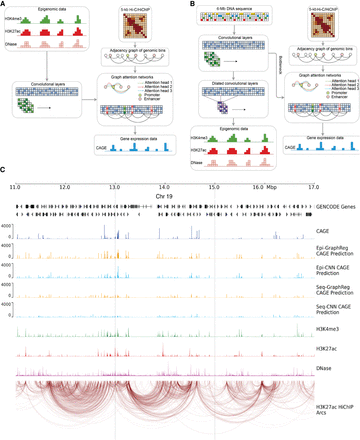
A schematic overview of GraphReg models. (A) The Epi-GraphReg model uses 1D epigenomic data, such as H3K4me3 and H3K27ac ChIP-seq and DNase-seq (or ATAC-seq) to learn local features of genomic bins via convolutional neural networks, and then propagates these features over adjacency graphs extracted from Hi-C/HiChIP contact matrices using graph attention networks to predict gene expression (CAGE-seq) across genomic bins. (B) The Seq-GraphReg model uses DNA sequence as input and, after some convolutional and dilated convolutional layers, predicts epigenomic data. This helps to learn useful latent representations of genomic DNA sequences that are then passed to the graph attention networks to be integrated over the adjacency graphs derived from Hi-C/HiChIP contact matrices and to predict gene expression values (CAGE-seq). (C) A 6-Mb genomic region (11 Mb–17 Mb) of Chr 19 showing input and output signals and predictions in K562 cells, including epigenomic data (H3K4me3, H3K27ac, DNase), CAGE, HiChIP interaction graph, and predicted CAGE values for GraphReg and CNN models. Training and evaluations of the models are performed in the dashed middle 2 Mb (here 13 Mb–15 Mb) region so that all genes can see the effects of their distal enhancers up to 2 Mb.











