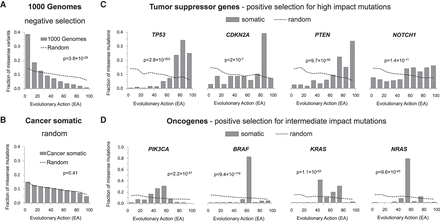
Coding variants under selection have nonrandom cohort integrals. The cohort integrals for all germline variants in the 1000 Genomes Project (A) and all somatic mutations from The Cancer Genome Atlas (TCGA; B). The somatic TCGA mutations were also shown for the tumor-suppressor genes TP53, CDKN2A, PTEN, and NOTCH1 (C) and for the oncogenes PIK3CA, BRAF, KRAS, and NRAS (D). The dashed lines correspond to simulated random amino acid changes in all human genes (A,B) or the respective single gene (C,D). The P-values of cohort integral differences between the observed and simulated random mutations were calculated by the one-tailed two-sample Kolmogorov–Smirnov test.











