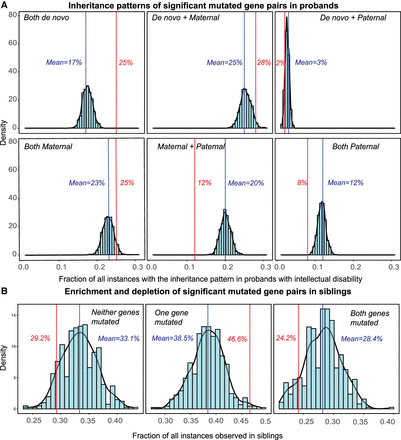
Analysis of parental and sibling inheritance patterns of significant gene pairs associated with ID. (A) Fraction of all instances of significant gene pairs observed within each of the six possible parental inheritance patterns (red) compared against 1000 simulations is shown (blue). During each simulation, random mutated gene pairs from the SSC cohort were selected, the inheritance status of the mutations was identified, and the fraction of those instances belonging to one of the six predefined categories was calculated. Comparing the observed fractions with the simulated fractions indicates statistical enrichment for two specific inheritance patterns based on empirical P-values: both variants being de novo, and one variant being de novo and the other transmitted from the mother. (B) Histograms show the carrier status of significant gene pairs in siblings of carrier probands (red) compared against 1000 simulations (blue). Among significant pairs, both genes were mutated in only 24.2% of all siblings (compared to 28.4% in simulations), whereas one of the two genes was mutated in 46.6% of all siblings (compared to 38.5% in simulations). These results show that mutations are more likely to be observed in just one of the two genes within the gene pairs and are less likely to be observed simultaneously in siblings of carrier probands.











