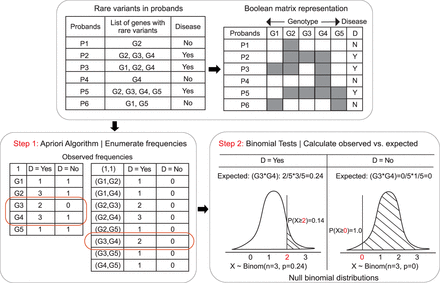
Conceptual overview of combinatorial analyses using RareComb. A Boolean representation of genotype (mutated genes, G1, G2, etc.) and disease status for probands (P1, P2, etc.) is shown. In step 1, the Apriori algorithm is applied to the Boolean input matrix to calculate the frequencies of individual (e.g., G1) and simultaneous occurrences of events (G1 and G2) that meet the user-specified criteria, including the size of combinations (pairs, triplets, etc.) and minimum frequency threshold of simultaneous occurrences. In step 2, independently in case and control groups, for each combination, the binomial test is applied to compare the observed frequency of simultaneous occurrence of events with its corresponding null binomial distribution of the expected frequencies calculated under the assumption of independence. Binomial test for gene pair G3 and G4 is shown as an example.











