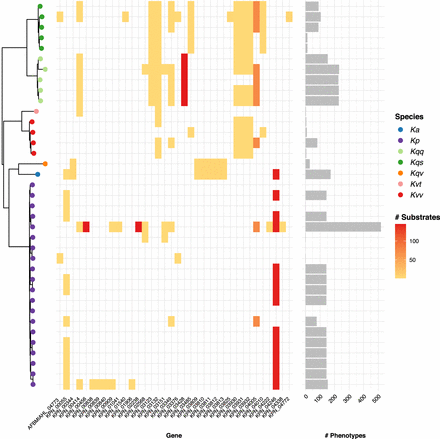
Variable loss-of-growth phenotypes. (Left) Core gene phylogeny per Figure 3, with tips colored by species as indicated in legend: (Ka) K. africana; (Kp) K. pneumoniae; (Kqq) K. quasipneumoniae subsp. quasipneumoniae; (Kqs) K. quasipneumoniae subsp. similipneumoniae; (Kqv) K. quasivariicola; (Kvt) K. variicola subsp. tropica; (Kvv) K. variicola subsp. variicola. (Middle) Heatmap showing core genes for which variable loss-of-growth phenotypes were predicted (columns). Shading indicates the number of substrates where loss of growth was predicted for each strain (rows) per the scale legend. (Right) Bars show the total number of loss-of-growth phenotypes predicted for each strain.











