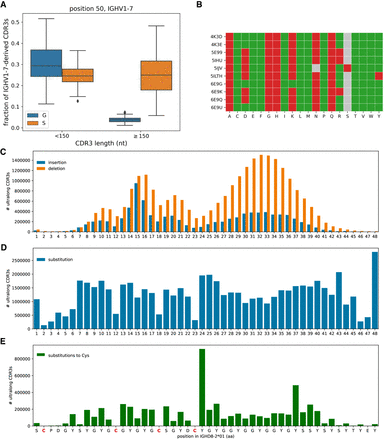
The anatomy of ultralong CDR3s. (A) Fractions of nonultralong CDR3s (lengths <150 nt) and ultralong CDR3s derived from IGHV1-7 with amino acids Gly (blue) and Ser (orange) at amino acid position 50. Fractions are computed in the combined data sets. (B) Impact of substitutions at position 50 in 10 crystallized antibodies predicted by the I-Mutant2.0 tool. Accession IDs of antibody structures are shown on the left. Gray cells show amino acids at position 50 present in structures (N or S). A green (red) cell (Ab, AA) indicates that mutation to amino acid AA is predicted to increase (decrease) stability of antibody structure Ab. (C–E) IGHD8-2 is a template for generating cysteines through SHMs. The germline sequence of IGHD8-2*01 is shown at the bottom with four cysteines shown in red. (C) The bar plot shows the number of ultralong CDR3s that have insertions (blue) and deletions (orange) at a given position of IGHD8-2*01. The position of insertion is defined as the position in the germline sequence that precedes the insertion. The average lengths of insertions and deletions in IGHD8-2 are 1.6 and 2.4 aa, respectively. (D) The bar plot shows the number of ultralong CDR3s that have a substitution at a given position of IGHD8-2*01. (E) The bar plot shows the number of ultralong CDR3s that have a substitution into cysteine at a given position of IGHD8-2*01.











