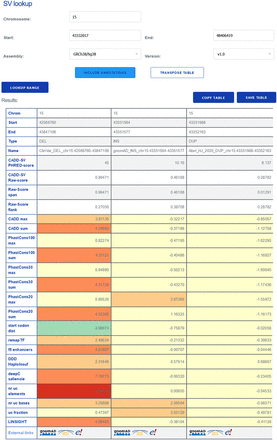
The CADD-SV web server can score custom SV sets, but it can also be used for direct lookup of prescored deletions, duplications, and insertions from gnomAD and ClinVar, as well as call-sets from Abel et al. (2020) and Beyter et al. (2021). For a given SV, the website provides the combined model scores as well as annotation values normalized to the range in the healthy gnomAD cohort (Z-score). This enables users to identify interesting variants from color-highlighted extreme feature values and not just by the combined CADD-SV score. Further, the website provides direct links for each SV to external resources like gnomAD, Ensembl, or the UCSC Genome Browser.











