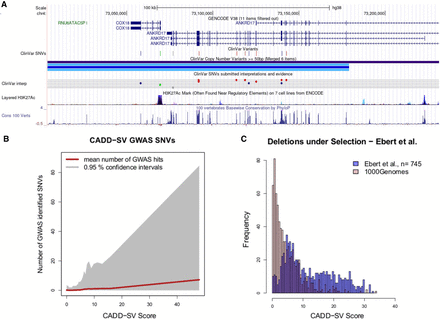
Prioritizing functional variants with CADD-SV. (A) Screenshot of UCSC Genome Browser tracks of a region (Chr 4: 73,004,055–73,231,324) deleted in one individual present in the gnomAD-SV cohort. Two genes are affected, with ANKRD17 variants being reported as causal for the autosomal dominant Chopra-Amiel-Gordon syndrome (CAGS). Various pathogenic SNVs were identified within the gene body of ANKRD17 and are marked in red in the UCSC ClinVar track. CAGS patients are characterized by developmental delay and moderate to severe intellectual disability. Further, various positions of this SV are highly conserved among 100 vertebrate genomes, contributing to CADD-SV's power of ranking it as a putatively deleterious variant. (B) Phred-scaled CADD-SV score distribution as a function of number of genome-wide association study-identified SNVs per deletion from gnomAD-SV. Especially among high scoring SVs, the average number of GWAS-associated SNVs increases drastically, suggesting functional variants in the pathogenic tail of the CADD-SV score distribution. (C) Scoring deletions under natural selection from Ebert et al. (2021). Shown are score distributions for the functional set (blue) against the same number of randomly drawn SVs from the 1000 Genomes Project. Note that we report Phred-scaled CADD-SV scores (log10 scale) with high values corresponding to high deleteriousness.











