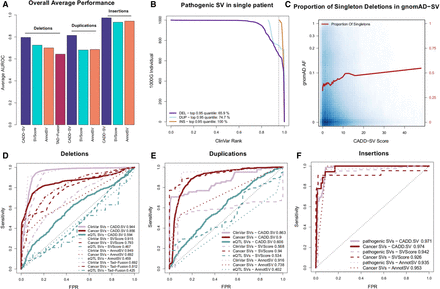
Validation set performance of the random forest models. (A) Summary of the performance of CADD-SV scores compared to SVScore, AnnotSV, and TAD-fusion scores across three validation sets (pathogenic variants, cancer variants, and putative eQTL SVs) for deletions, duplications, and insertions. (B) Rank of ClinVar pathogenic SVs added to SVs of healthy individuals from the 1000 Genomes Project. CADD-SV prioritizes the pathogenic SVs over the other SVs in a single simulated patient, scoring pathogenic variants in the top fifth percentile of deletions, duplications, and insertions for 65.9%, 74.7%, and 100% of simulated variant sets, respectively. (C) CADD-SV score distribution as a function of gnomAD allele frequency. Higher CADD-SV values represent an increased likelihood to be deleterious. In the deleterious tail of the score distribution, there is an excess of singletons (shown in red; bin size 0.025), which hints at negative selection against deleterious deletions. (D–F) CADD-SV performance of various validation sets compared to common gnomAD SVs (AF ≥ 0.05). Performance is measured as sensitivity over false positive rate. CADD-SV is able to identify ClinVar pathogenic SVs (n = 3262 deletions, 82 duplications, and 78 insertions, pale red) as well as SVs reported in the ICGC cancer cohort (n = 52,677 deletions, 42,972 duplications, and 18 insertions, dark red) from common SVs in gnomAD. Further, CADD-SV can identify noncoding SVs that are associated with differences in gene expression (turquoise). CADD-SV scores (solid lines) are compared to SVScore (dashed lines), AnnotSV (dotted lines), and TAD-fusion (dashed and dotted lines) for deletions (D), duplications (E), and insertions (F).











