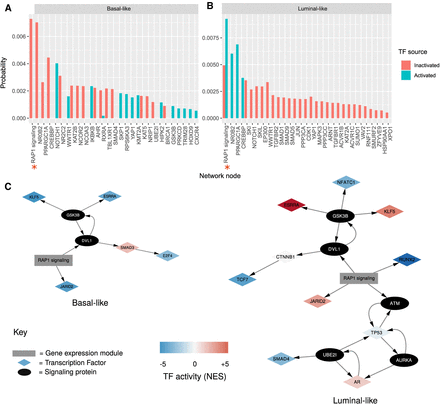
Network propagation of active TFs within cell-shape network. (A,B) Bar plot showing network propagation in a predicted cell-shape network from activated and inactivated TFs in basal-like cell lines (A) and luminal (B). The y-axis is a steady-state probability (or the “heat” of the nodes in the network after the diffusion) over the graph imposed by the starting seeds, ordered by size. Red bars represent propagation from TF seeds that are predicted to be inactivated, and blue bars show propagation from TF seeds that are predicted to be activated. Red asterisks along the x-axis indicate supernodes that represent gene expression modules. Only those nodes with combined probability > 0.0001 are shown, with the full results available in Supplemental Table S9. (C) Subnetworks illustrating the paths between activated TFs (in basal-like and luminal-like) and the “RAP1 signaling” gene expression module. TFs are shown as diamond-shaped nodes, with their color representing their activity. The “RAP1 signaling” gene expression module is shown as a gray rectangle. Signaling proteins are shown as black nodes.











