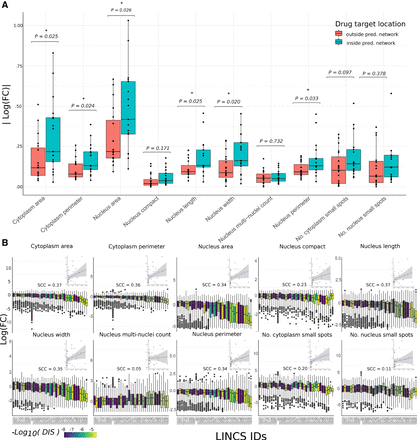
Effect of drug perturbation of derived network on breast cancer cell line morphology. (A) Box plots showing the absolute log10 fold changes after treatment with a drug relative to a control for each cell-shape variable. The drugs are grouped by those targeting kinases within the predicted regulatory network (blue) and those targeting other kinases not predicted to be associated with cell shape (red). P-values (Welch two-sample t-test) are shown with asterisks indicating significance. (B) Bar plot showing the absolute difference in log fold changes of cell-shape variables after treatment with a drug relative to a control. Here, each drug is shown separately (with the LINCs ID shown on the x-axis) and colored based on the drug influence score (DIS), and each data point represents a single cell. Insets are plots showing the correlation between this influence score and the difference between mean treated cells and mean control cells in each of the 10 measured cell-shape features for each drug. Spearman's correlation coefficients are shown above the inset plots.











