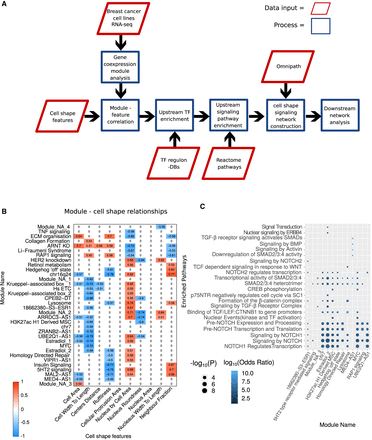
Overview of workflow and resultant gene expression modules and pathways. (A) Schematic illustrating the steps involved in phenotype-specific network construction. Gene expression modules are identified by integrating cell-shape variables (derived from imaging data) with RNA-seq data from breast cancer cell lines. These gene expression modules are correlated with specific cell-shape features to find morphologically relevant modules. Next, transcription factors (TFs) are identified whose targets significantly overlap with the contents of the expression modules. These TFs are used to identify pathways regulating the gene expression modules, which are then integrated to form a contiguous network using PCSF. (B) Heat map of significantly correlated gene expression module eigengenes with cell-shape features. Non significant interactions were set to zero for clarity. (C) Dot plot illustrating the enrichment of pathways among TFs found to regulate gene expression modules. The x-axis shows the module names (as defined by Supplemental Table S3), and the y-axis shows the signaling pathways found to be significantly (P < 0.01) enriched in the TFs that regulate the given module (as defined by Supplemental Table S5). The y-axis is arranged such that the terms with the highest combined odds ratio are at the bottom. Size of the dot represents the −log10(P), and the color indicates a log10 transformation of the odds ratio.











