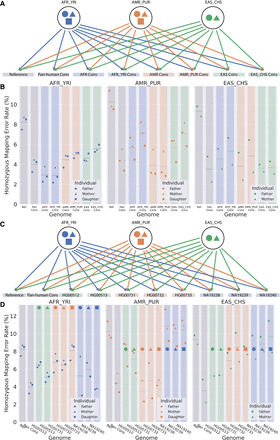
Mapping accuracy improvements owing to switching from reference to consensus genomes when mapping to alternative genomes. (A) Each individual from each population is independently mapped to the reference, pan-human consensus, and all population and superpopulation consensus genomes. (B) Homozygous mapping error rate when mapping to different consensus. The color of the marker indicates the population to which that individual belongs, whereas the shape of the marker identifies the individual within the trio. The color of the background rectangle indicates the population of the genome. The dashed line in each column represents the mean mapping error for that combination of genome and individuals. (C) Each individual from each population is independently mapped to the reference, pan-human consensus, and all personal haploid genomes. (D) Homozygous mapping error rate when mapping to different personal haploid genomes.











