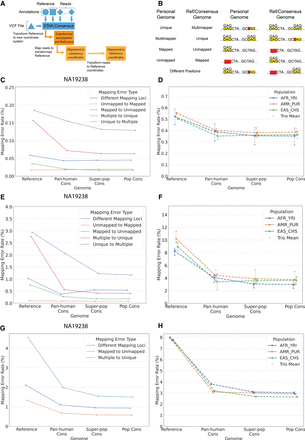
Mapping accuracy improvements owing to switching from reference to consensus genomes. (A) Internal workflow of STAR-consensus. (B) Different types of mapping errors based on the read's mapping status in the individual's haploid personal genome and the reference or given consensus genome. (C) Overall mapping error rate for each error type for individual NA19238. Genome is shown on the x-axis, and the mapping error rate is shown on the y-axis. (D) Overall mapping error rate for all individuals. Individuals from the same population are grouped together by color, with each marker shape representing one individual in the population. The dashed line shows the average error rate for the population, and the solid vertical line indicates the range of the population. (E) Homozygous mapping error rate for each error type for individual NA19238. (F) Homozygous mapping error rate for all individuals. Individuals from the same population are grouped together by color, with each marker shape representing one individual in the population. The dashed line shows the average error rate for the population, and the solid vertical line indicates the range of the population. (G) Homozygous mapping error rates for each error type for simulated reads for individual NA12938. (H) Population-average homozygous mapping error rates for simulated reads for all individuals.











