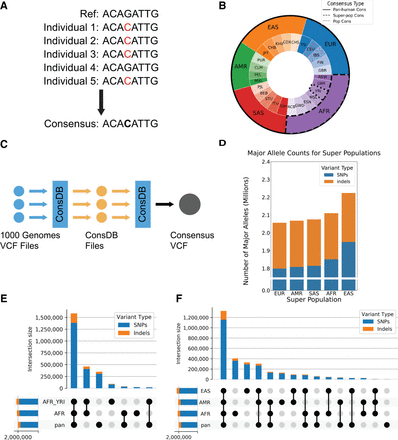
Construction of the consensus genome with major allele replacements. (A) Construction of a consensus genome: The minor allele in the reference is replaced by the most common (major) allele in the population. (B) Visual representation of the individuals used to construct consensus genomes of varying population specificity. (C) ConsDB workflow. (D) Number of major alleles for each population consensus genome that were replaced in the reference. (E) Number of SNPs and indels shared between different combinations of the pan-human, superpopulation, and population consensus genomes for the African population. The bars in the top bar plot show the number of SNPs and indels that are unique to the intersection of genomes indicated in the dot matrix below. The horizontal bars on the bottom left show the total number of SNPs and indels present in each genome. (F) Number of SNPs and indels shared between different combinations of the pan-human consensus and all three superpopulation consensus genomes. The bars in the top bar plot show the number of SNPs and indels that are unique to the intersection of genomes indicated in the dot matrix below. The horizontal bars on the bottom left show the total number of SNPs and indels present in each genome.











