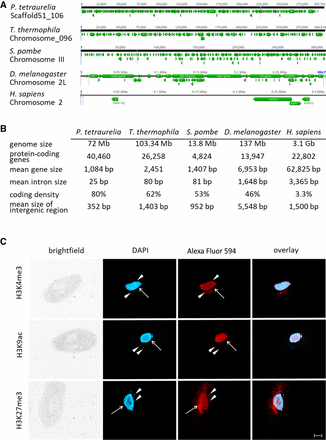
Features of the Paramecium genome in comparison to other organisms. (A) Comparisons of distribution of genes (green arrows) along the chromosomes of selected organisms to highlight the variation in coding density (P. tetraurelia, Tetrahymena thermophila, Schizosaccharomyces pombe, Drosophila menlanogaster, Homo sapiens). A window of 300 kb is shown for each chromosome in a genome browser. (B) Summary of genomic features of the same organisms named in A. For details on collected data, see Methods. (C) Detection of histone modifications in vegetative Paramecium nuclei by immunofluorescence staining. DNA in the nuclei is stained with DAPI (blue), and antibodies directed against the three indicated modifications (H3K4me3, H3K9ac, H3K27me3) were labeled with a secondary Alexa Fluor 594 conjugated antibody (red). Arrowheads point at micronuclei; arrows indicate position of the macronucleus. Other panels show brightfield and overlay of signals. Representative overlays of Z-stacks of magnified views are shown. Scale bar, 10 µm.











