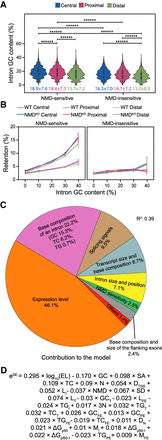
GC content related to nucleosome positioning contributes to intron splicing efficiency. (A) GC content (%) distribution of introns based on the distance to the closest nucleosome center and NMD sensitivity. Mean and standard deviation for each group is reported at the bottom. P-values were calculated using the Mann–Whitney U test and adjusted using the false discovery rate (5%). Tests were run between introns belonging to the same positional group or between introns belonging to the same NMD group. (*) P < 0.05, (**) P < 10−2, (***) P < 10−3, (****) P < 10−4, (*****) P < 10−5, (******) P < 10−6. (B) The retention rate of introns in WT and NMD-depleted (NMDKD) cells as a function of their GC content (excluded GT and AG dinucleotides at both extremities). Introns are classified based on their distance to the closest nucleosome center and on whether they are NMD sensitive or not. Binning = 10%. Error bars represent the SEM. P-values calculated using the Mann–Whitney U test, and adjusted using a false-discovery rate (5%), are displayed in Supplemental Figure S4A. (C) Modeling splicing efficiency (SE) in NMD-depleted cells: The pie chart reports the contribution of each parameter or group of parameters used in the final model. The full list of retained parameters, reporting their contribution and their statistical significance, is displayed in Supplemental Table S1 as well as in Supplemental Figure S4C. (D) The full fitted model in explaining intron splicing efficiency, indicating whether each parameter is positively or negatively correlated with splicing efficiency. The parameter abbreviations are explained in Supplemental Table S1.











