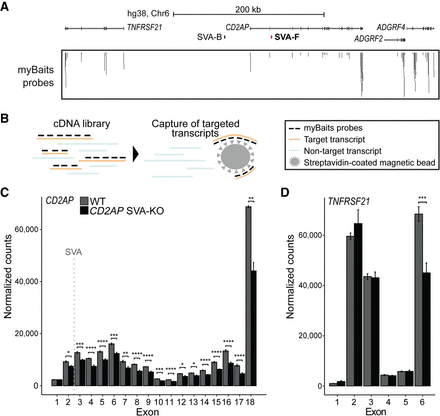
Intronic SVA deletion alters exon expression of CD2AP gene. (A) Overview of CD2AP locus. Location of SVA removed by CRISPR-Cas9 KO shown in red. Probes from myBaits targeting CD2AP exons. (B) Schematic of capture of targeted transcripts with myBaits probes. (C,D) Normalized mean expression of three replicates shown per exon of CD2AP and the nearby highly expressed gene TNFRSF21. Location of SVA indicated with dashed gray line. Adjusted P-value from DESeq2 shown for each exon. (****) P < 0.0001, (***) P < 0.001, (**) P < 0.01, (*) P < 0.05.











