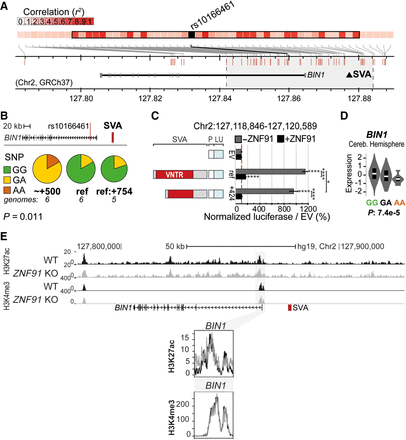
Structurally variable SVA near BIN1 links to a disease-associated SNP and has the potential to differentially regulate nearby genes. (A) Overview of LD block for rs10166461, with area r2 > 0.8 highlighted in gray. Approximate location of SVA marked with black triangle. (B) rs10166461 genotyping analysis for individuals homozygotic for BIN1-SVA variants ref (n = 6) and +424 or +521 (∼+500, n = 6) and heterozygotic for ref and +754 (n = 5). Ancestral allele = G, risk allele = A. Fisher's exact test: P = 0.0108. (C) Schematic overview of luciferase constructs (P = minimal promoter, LU = luciferase gene) with BIN1-SVA variants, with corresponding luciferase activity in transfected mESCs with and without ZNF91. Two-way ANOVA with Tukey's multiple comparison. (****) P < 0.0001, (*) P < 0.05. Error bars: SEM. (D) Analysis of eQTL data in cerebellar hemisphere for rs10166461 and BIN1. (E) KO of the SVA repressor ZNF91 does not influence H3K27ac and H3K4me3 at the promoter of BIN1 in hESCs. (Top) Overview of locus, (bottom) magnification of regions of interest.











