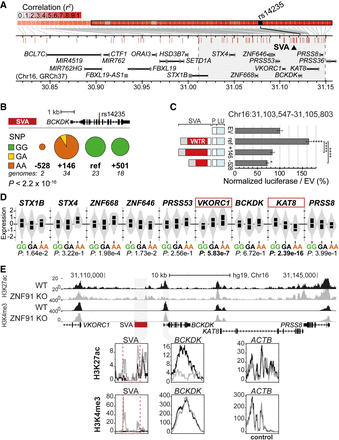
Structurally variable SVA near BCKDK links to a disease-associated SNP and has the potential to differentially regulate nearby genes. (A) Overview of LD block for rs14235, with area r2> 0.8 highlighted in gray. Approximate location of SVA marked with black triangle. (B) rs14235 genotyping analysis for individuals homozygotic for BCKDK-SVA variants −600 (n = 2), ref (n = 23), +150 (n = 34), and +500 (n = 18). (Ancestral allele) G; (risk allele) A. Fisher's exact test: P < 2.2 × 10−16. (C) Schematic overview of luciferase constructs (P = minimal promoter, LU = luciferase gene) with BCKDK-SVA variants (Chr 16: 31,103,547–31,105,803 GRCh38), with corresponding luciferase activity in transfected mESCs. N3n9, except BCKDK-SVA ref (n = 8). One-way ANOVA with Tukey's multiple comparison, (****) P < 0.0001, (*) P < 0.05. Error bars: SEM. (D) Analysis of eQTL data in cortex for rs14235 for genes within the LD block with r2 > 0.8. Normalized expression is shown. Genes considered significant are shown in a red box. (E) KO of the SVA repressor ZNF91 lowers H3K27ac at the promoter of BCKDK and increases H3K4me3 methylation at the SVA near BCKDK in hESCs. ACTB shown as control enhancer region. (Top) Overview of locus, (bottom) magnification of regions of interest.











